Get PeerJ Article Alerts

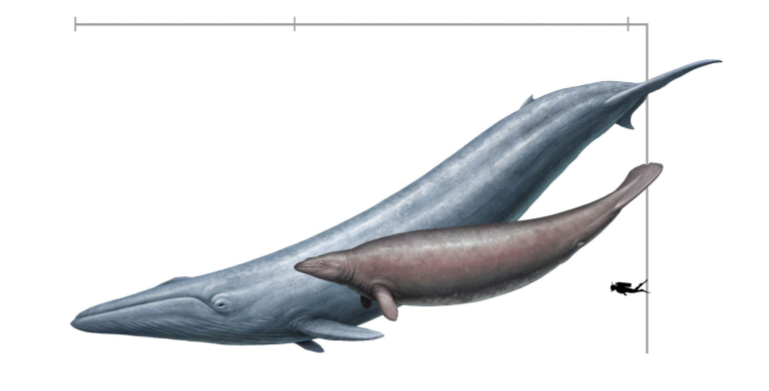
Article Spotlight: The dinosaurs that weren’t: osteohistology supports giant ichthyosaur affinity of enigmatic large bone segments from the European Rhaetian
April 12, 2024

Article Spotlight: The role of temperature on the development of circadian rhythms in honey bee workers
April 9, 2024